Dear brew enthusiasts
In this weeks episode, we will focus on the archaeology of beer brewing. I am sure you have read about the recent discovery of a large brewery in ancient Egypt. Now, few will question me when I say that this is an exciting discovery. However, I will not talk about it since it has received enough coverage on the web. It did inspire me to look at other archaeological research on brewing. Perhaps, you will not be surprised to learn that genome sequencing and computational analyses have helped travel back in time and answer some interesting questions.
Here, I will discuss a paper by the Christian Landry group that describes the isolation, characterization and genome sequencing of a yeast strain found in an archaeological site in Québec, Canada. The authors could catch living yeast cells roaming in known old brewing facilities through a basic sampling approach.
Genome sequencing of this strain (termed "Jean-Talon) and comparisons to sequences from modern beer yeasts helped the authors establish the probable origin of the yeast strange used in the late 19th century. Growth assays further helped established the strains provenance, suggesting that while it originates in Europe, it may have been used as late as the 1960s
Let's dive in.
To isolate strains, the authors sampled the vaults of the Intendant's Palace of Nouvelle France. The building is a historical site in Québec City, where brewing started in the early 17th until the 20th century when cobalt poisoning halted beer production. Media plates were left open in the vaults and incubated to grow yeast colonies for characterization.
The authors captured a series of yeast colonies this way, most of which turned out to be Saccharomyces cerevisiae. DNA staining, followed by quantification, then helped establish that these cells' ploidy level is 4n (4 copies of each chromosome). Representative strains were then sequenced, generating a view of genetic diversity within the genome.
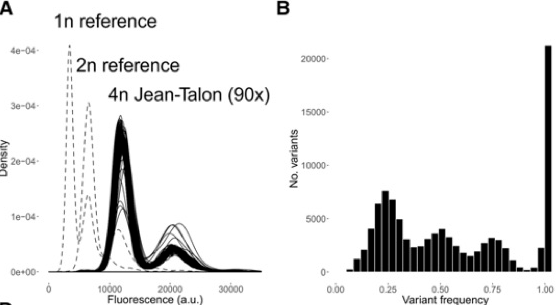
Figure 2 from Fijarczyk et al (2020). Principal Component analyses of Jean-Talon vs collections of known yeast strains. By using SNP data from two distinct datasets, the authors establish that the JT strain is closely related to strains used in beer and bread making.
Having obtained the genome sequence, one can look for single nucleotide polymorphisms. These are single nucleotide sequence variations that scientists can use to study genetic diversity and relatedness between strains. By comparing the set of SNPs in the Jean-Talon strain and comparing this with two previously generated datasets, describing a broader collection of strains, the authors were able to place their strain into a group of isolates, known to be used for beer and bread making. Furthermore, their analyses indicate that Jean-Talon is most related to strains that have come from Europe rather than North America.
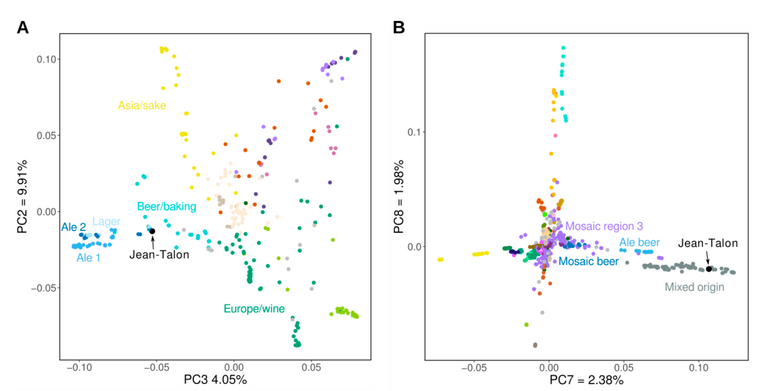
Figure 2 from Fijarczyk et al (2020). Principal Component analyses of Jean-Talon vs collections of known yeast strains. By using SNP data from two distinct datasets, the authors establish that the JT strain is closely related to strains used in beer and bread making.
Of course, these findings raise questions about when the strain was introduced or used for the first time. Does the strain originate from the early 17th century when colonists started the brewery? Or is Jean-Talon a contemporary yeast in use as late as the 1960s?
The answer is maybe both. While the strain appears to have European ancestry, yeast growth assays in the presence of Cobalt helped establish that the Jean-Talon strain is relatively tolerant of this element. Further analyses found that tolerance to Cobalt is higher than levels observed in many other yeast strains. These results suggest that Jean-Talon was used extensively on site where it was allowed to adapt to higher levels of Cobalt.
While the paper goes into much further detail on the evolution, organization and function of the Jean-Talon genome, I will leave it here and emphasize how a combination of genome sequencing, comparative analyses and functional assays can help us untangle the history of the living beings that surround us.
Cheers,
Edgar, The Beerologist
Edgar Huitema is a Scientist, Brewer & Scientific Consultant at ExtrAnalytics. Get in touch to learn about what we can do for you.
For the manuscript describing this work, Please find the reference below:
Fijarczyk, A., Hénault, M., Marsit, S., Charron, G., Fischborn, T., Nicole-Labrie, L., & Landry, C. R. (2020). The genome sequence of the Jean-Talon strain, an archeological beer yeast from Québec, reveals traces of adaptation to specific brewing conditions. G3: Genes, Genomes, Genetics, 10(9), 3087-3097.
Thanks for your contribution to the STEMsocial community. Feel free to join us on discord to get to know the rest of us!
Please consider supporting our funding proposal, approving our witness (@stem.witness) or delegating to the @stemsocial account (for some ROI).
Please consider using the STEMsocial app app and including @stemsocial as a beneficiary to get a stronger support.